1.Kết nối dữ liệu data 2010-2016 - remove duplicate
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
# drug1016.Edited %>% filter(Group=="AntibacterialSystematic") %>% filter(ATC=="")
# 0 rows -> OK no drug withou ATC code in antibiotic
betalactam1016=drug1016.Edited %>%mutate(GrPhar=substr(ATC,1,4),
Year=substr(as.character(Year.ID),1,4)) %>%
filter(Group=="AntibacterialSystematic") %>%
filter(GrPhar %in% c("J01D","J01C")) %>% unique()
id=betalactam1016$Year.ID
# #To return ALL MINUS ONE duplicated values:
# id[duplicated(id)]
#
# #To return ALL duplicated values by specifying fromLast argument:
# id[duplicated(id) | duplicated(id, fromLast=TRUE)]
#Yet another way to return ALL duplicated values, using %in% operator:
id.trung1016= unique(id[id %in% unique(id[duplicated(id)])])
2.List report combination with betalactamase
1
2
3
4
5
6
7
8
9
10
# elimination of coprescription 2 suspected betaclam
betalactam1016 %>% filter(Year.ID %in% id.trung1016) %>% View()
id.betalactamse1016=c( 201098,201096, 2010556, 20101100, 20101355, 20101374, 20101399, 20101401,
20101406, 20101438, 20101443, 20101457, 20101492
) %>% as.character()
betalactam1016 %>% filter(Year.ID %in% id.betalactamse1016) # loai do 2 betalactam tren 1 thuoc
1
2
3
4
5
6
7
8
9
10
11
12
13
14
## # A tibble: 27 x 6
## ATC Drug Year.ID Group GrPhar Year
## <chr> <chr> <dbl> <chr> <chr> <chr>
## 1 J01CA01 ampicillin 2e+05 AntibacterialSystematic J01C 2010
## 2 J01DD01 cefotaxim 2e+05 AntibacterialSystematic J01D 2010
## 3 J01CG01 sulbactam 2e+05 AntibacterialSystematic J01C 2010
## 4 J01CA01 ampicillin 2e+05 AntibacterialSystematic J01C 2010
## 5 J01CG01 sulbactam 2e+05 AntibacterialSystematic J01C 2010
## 6 J01CR acid clavulanic 2e+06 AntibacterialSystematic J01C 2010
## 7 J01CA04 amoxicillin 2e+06 AntibacterialSystematic J01C 2010
## 8 J01CR acid clavulanic 2e+07 AntibacterialSystematic J01C 2010
## 9 J01CA04 amoxicillin 2e+07 AntibacterialSystematic J01C 2010
## 10 J01CR acid clavulanic 2e+07 AntibacterialSystematic J01C 2010
## # ... with 17 more rows
1
id.betalactamse1016[-2] # loai gia tri thu 2 tuong duong bao cao 201096
1
2
## [1] "201098" "2010556" "20101100" "20101355" "20101374" "20101399"
## [7] "20101401" "20101406" "20101438" "20101443" "20101457" "20101492"
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
betalactam1016=betalactam1016 %>% mutate(
Drug=replace(Drug,Year.ID=="201098","ampicilin/sulbactam"),
Drug=replace(Drug,Year.ID=="2010556","amoxicilin/acid clavulanic"),
Drug=replace(Drug,Year.ID=="20101100","amoxicilin/acid clavulanic"),
Drug=replace(Drug,Year.ID=="20101355","amoxicilin/acid clavulanic"),
Drug=replace(Drug,Year.ID=="20101374","amoxicilin/acid clavulanic"),
Drug=replace(Drug,Year.ID=="20101399","amoxicilin/acid clavulanic"),
Drug=replace(Drug,Year.ID=="20101401","amoxicilin/acid clavulanic"),
Drug=replace(Drug,Year.ID=="20101406","amoxicilin/acid clavulanic"),
Drug=replace(Drug,Year.ID=="20101438","cefoperazon/sulbactam"),
Drug=replace(Drug,Year.ID=="20101443","amoxicilin/acid clavulanic"),
Drug=replace(Drug,Year.ID=="20101457","amoxicilin/acid clavulanic"),
Drug=replace(Drug,Year.ID=="20101492","piperacillin/tazobactam")) %>%
mutate(Drug=str_replace_all(Drug,"cilin","cillin")) %>%
select(-ATC) %>% unique()
# betalactam1016 %>% View
id=betalactam1016$Year.ID
ds.trung.final1016=betalactam1016 %>% filter(Year.ID %in% id[id %in% unique(id[duplicated(id)])],
!Year.ID %in% id.betalactamse[-2])%>% unique() # 181
betalactam.nodup1016=betalactam1016%>% filter(!Year.ID%in%ds.trung.final$Year.ID) %>% unique()
betalactam.nodup.casenoncase.1016=betalactam.nodup1016 %>%left_join(
d1016.2 %>% select(Year,Year.ID=mabc,Age,Sex,Case,Mortality)
) %>%
mutate( Drug=replace(Drug,Year.ID=="20147153","imipenem/cilastatin")) %>% filter(Case %in% 0:1) %>%
mutate(
Drug=replace(Drug,Drug=="amipicillin/sulbactam","ampicillin/sulbactam"),
Drug=replace(Drug,Drug=="cefotaxime","cefotaxim"),
Drug=replace(Drug,Drug=="cefpirome","cefpirom"),
Drug=replace(Drug,Drug=="ceftriaxone","ceftriaxon"),
Drug=replace(Drug,Drug=="amoxicillin/clavunanic","amoxicillin/acid clavulanic"),
Drug=replace(Drug,Drug=="benzathin penicillin g","benzathine benzylpenicillin"),
Drug=replace(Drug,Drug=="penicillin g","benzylpenicillin"),
Drug=replace(Drug,Drug=="penicillin v","phenoxymethylpenicillin")
)%>% left_join(read_excel("Betalactam ATC code update.xlsx") %>% select(Drug,ATC)) %>%
mutate(subgroup.betalactam=substr(ATC,1,5))
1
betalactam.nodup.casenoncase.1016 %>% group_by(Drug) %>% tally(sort=T) %>% unique()
1
2
3
4
5
6
7
8
9
10
11
12
13
14
## # A tibble: 54 x 2
## Drug n
## <chr> <int>
## 1 cefotaxim 3115
## 2 ceftriaxon 1561
## 3 ceftazidim 1192
## 4 cefuroxim 703
## 5 amoxicillin 701
## 6 cefoperazon 405
## 7 cefalexin 371
## 8 ampicillin 278
## 9 amoxicillin/acid clavulanic 271
## 10 ampicillin/sulbactam 193
## # ... with 44 more rows
1
betalactam.nodup.casenoncase.1016 %>% group_by(GrPhar) %>% tally(sort=T) %>% unique()
1
2
3
4
5
## # A tibble: 2 x 2
## GrPhar n
## <chr> <int>
## 1 J01D 8892
## 2 J01C 1904
1
betalactam.nodup.casenoncase.1016 %>% group_by(subgroup.betalactam) %>% tally(sort=T) %>% unique()
1
2
3
4
5
6
7
8
9
10
11
12
13
14
## # A tibble: 11 x 2
## subgroup.betalactam n
## <chr> <int>
## 1 J01DD 6824
## 2 J01CA 1054
## 3 J01DC 975
## 4 J01DB 735
## 5 J01CR 614
## 6 J01DE 220
## 7 J01CE 160
## 8 J01DH 138
## 9 J01CF 74
## 10 J01CG 1
## 11 J01D 1
3.Subgroup signals : J01DD and J01CE
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
subgroup.betalactam.list=betalactam.nodup.casenoncase.1016%>%group_by(subgroup.betalactam) %>%
tally(sort=T) %>% filter(subgroup.betalactam!="J01D") #%>% View ()
for (i in 1:nrow(subgroup.betalactam.list)){
temp=betalactam.nodup.casenoncase.1016 %>% filter(., grepl(subgroup.betalactam.list[i,]$subgroup.betalactam, subgroup.betalactam))
varname <- paste(subgroup.betalactam.list[i,]$subgroup.betalactam )
betalactam.nodup.casenoncase.1016[[varname]]=with(betalactam.nodup.casenoncase.1016,if_else(Year.ID%in% temp$Year.ID,1,0))
}
OR.subgroup.betalactam=NULL
for (i in 1:nrow(subgroup.betalactam.list)){
formula.i=lapply(subgroup.betalactam.list[i,]$subgroup.betalactam,
function(var) formula(paste0('Case~',var),env=globalenv()))
OR.subgroup.betalactam=rbind(OR.subgroup.betalactam,
betalactam.nodup.casenoncase.1016 %>%
select(Year,Age,Sex,Case,matches(subgroup.betalactam.list[i,]$subgroup.betalactam)) %>%
glm(formula=formula.i[[1]],family=binomial(link="logit")) %>% OR95()%>%
tibble::rownames_to_column() %>% filter(between(row_number(), 2, n()))
)
}
OR.subgroup.betalactam.n=betalactam.nodup.casenoncase.1016 %>% filter(Year.ID %in% subset(betalactam.nodup.casenoncase.1016,Case==1)$Year.ID) %>% group_by(subgroup.betalactam) %>% tally(sort=T) %>% left_join(
OR.subgroup.betalactam , by = c("subgroup.betalactam" = "rowname")
)
OR.subgroup.betalactam.n %>% print()
1
2
3
4
5
6
7
8
9
10
11
12
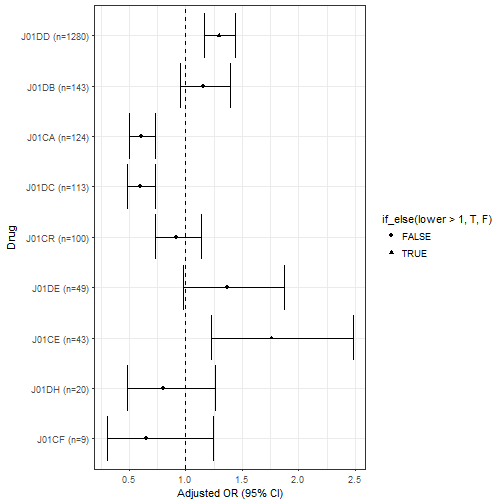
## # A tibble: 9 x 5
## subgroup.betalactam n OR lower upper
## <chr> <int> <dbl> <dbl> <dbl>
## 1 J01DD 1280 1.30 1.17 1.44
## 2 J01DB 143 1.16 0.95 1.39
## 3 J01CA 124 0.61 0.50 0.73
## 4 J01DC 113 0.60 0.49 0.73
## 5 J01CR 100 0.92 0.73 1.14
## 6 J01DE 49 1.37 0.98 1.87
## 7 J01CE 43 1.76 1.22 2.48
## 8 J01DH 20 0.80 0.48 1.26
## 9 J01CF 9 0.65 0.30 1.25
1
2
3
4
5
6
7
8
9
10
OR.subgroup.betalactam.n %>% unique() %>%
mutate(subgroup.betalactam=paste0(subgroup.betalactam,' (n=',n,")")) %>%
arrange(n) %>% mutate(subgroup.betalactam = factor(subgroup.betalactam, subgroup.betalactam)) %>%
ggplot( aes(x=subgroup.betalactam, y=OR, ymin=lower, ymax=upper)) +
geom_errorbar() +
geom_point(aes(shape=if_else(lower>1,T,F))) + #,col="blue",shape=5
geom_hline(yintercept=1, lty=2) + # add a dotted line at x=1 after flip
coord_flip() + # flip coordinates (puts labels on y axis)
xlab("Drug") + ylab("Adjusted OR (95% CI)") +
theme_bw() # use a white background
«««< HEAD:_posts/2018-05-30-Betalactam analysis stratification Notebook.md
 =======
=======

1522bad2ad157ed42d6809c938b743194108ad6a:_posts/2018-05-30-Betalactam analysis stratification Notebook.md
3.1.Signal of subgroup.cepha
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
for (i in 1:length(subgroup.cepha)){
temp=cepha %>% filter(., grepl(subgroup.cepha[i], cephagroup))
varname <- paste(subgroup.cepha[i] )
d1016.3[[varname]]=with(d1016.3,if_else(mabc%in% temp$Year.ID,1,0))
}
OR.table.subgroupcepha.1016=NULL
for (i in 1:length(subgroup.cepha)){
formula.i=lapply(subgroup.cepha[i],
function(var) formula(paste0('Case~',var),env=globalenv()))
OR.table.subgroupcepha.1016=rbind(OR.table.subgroupcepha.1016,
d1016.3 %>% #filter(ATC!="J01D") %>%
select(Year,Age,Sex,Case,matches(subgroup.cepha[i])) %>%
glm(formula=formula.i[[1]],family=binomial(link="logit")) %>% OR95()%>%
tibble::rownames_to_column() %>% filter(between(row_number(), 2, n()))
)
}
OR.table.subgroupcepha.1016 %>% print()
1
2
3
4
5
6
## rowname OR lower upper
## 1 J01DD 2.3 2.12 2.4
## 2 J01DB 1.8 1.52 2.1
## 3 J01DC 1.1 0.95 1.3
## 4 J01DH 1.1 0.76 1.6
## 5 J01DE 2.3 1.80 3.0
1
2
3
4
5
6
7
8
9
10
11
12
13
14
OR.table.subgroupcepha.1016.n=cepha %>% left_join(d1016.3 %>% select(Year.ID=mabc,Case) %>% filter(Case==1))%>% filter(Case==1) %>% group_by(cephagroup) %>% tally() %>% filter(cephagroup!="J01D") %>%
left_join(OR.table.subgroupcepha.1016 ,by = c("cephagroup" = "rowname"))
OR.table.subgroupcepha.1016.n %>% unique() %>% mutate(cephagroup=paste0(cephagroup,'(n=',n,")")) %>%
arrange(n) %>% mutate(cephagroup = factor(cephagroup, cephagroup)) %>%
ggplot( aes(x=cephagroup, y=OR, ymin=lower, ymax=upper)) +
geom_errorbar() +
geom_point(aes(shape=if_else(lower>1,T,F))) + #,col="blue",shape=5
geom_hline(yintercept=1, lty=2) + # add a dotted line at x=1 after flip
coord_flip() + # flip coordinates (puts labels on y axis)
xlab("cephagroup") + ylab("Adjusted ROR (95% CI)") +
theme_bw() # use a white background

1
2
3
4
5
6
OR.table.subgroupcepha.1016.n %>%
arrange(desc(n)) %>% mutate(OR=round(OR,2),
lower=round(lower,2),
upper=round(upper,2)) %>%
write.csv2("OR.table.subgroupcepha.1016.n.csv",row.names=F)
4.C3G vs other cephalosporins: subgroup.betalactam
1
2
3
4
5
6
7
8
9
10
11
x=betalactam.nodup.casenoncase.1016$Drug
x=x %>% stringr::str_replace_all(c(" "),".") %>%
stringr::str_replace_all(c("/"),".")
betalactam.nodup.casenoncase.1016$Drug=x
cepha.strata1=betalactam.nodup.casenoncase.1016 %>% filter(GrPhar=="J01D")
# mutate(cephagroup=substr(ATC,1,5)) %>%
# filter(ATC!="J01D")
cepha.strata1 %>%group_by(subgroup.betalactam) %>% tally(sort=T) %>% print()
1
2
3
4
5
6
7
8
9
## # A tibble: 6 x 2
## subgroup.betalactam n
## <chr> <int>
## 1 J01DD 6823
## 2 J01DC 975
## 3 J01DB 735
## 4 J01DE 220
## 5 J01DH 138
## 6 J01D 1
1
2
3
4
5
6
cepha.strata.C3G=betalactam.nodup.casenoncase.1016 %>% filter(subgroup.betalactam=="J01DD")
d1016.strata.C3G=d1016.3 %>% select(Year,Year.ID=mabc,Age,Sex,Mortality,Case) %>%
filter(Year.ID%in%cepha.strata.C3G$Year.ID)
cepha.strata.C3G.list=cepha.strata.C3G %>% group_by(Drug) %>% tally(sort=T)
5 Strata cepha third geneation : d1016.strata.C3G
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
d1016.strata.C3G[["C3G"]]=""
for (i in 1:nrow(cepha.strata.C3G.list)){
temp=cepha.strata.C3G %>% filter(., grepl(cepha.strata.C3G.list[i,]$Drug, Drug))
varname <- paste(cepha.strata.C3G.list[i,]$Drug )
d1016.strata.C3G[[varname]]=with(d1016.strata.C3G,if_else(Year.ID%in% temp$Year.ID,1,0))
d1016.strata.C3G=d1016.strata.C3G %>% mutate(C3G=if_else(Year.ID%in% temp$Year.ID,varname,C3G))
}
OR.cepha.strata.C3G=NULL
for (i in 1:nrow(cepha.strata.C3G.list)){
formula.i=lapply(cepha.strata.C3G.list[i,]$Drug,
function(var) formula(paste0('Case~',var),env=globalenv()))
OR.cepha.strata.C3G=rbind(OR.cepha.strata.C3G,
d1016.strata.C3G %>%
select(Year,Age,Sex,Case,matches(cepha.strata.C3G.list[i,]$Drug)) %>%
glm(formula=formula.i[[1]],family=binomial(link="logit")) %>% OR95()%>%
tibble::rownames_to_column() %>% filter(between(row_number(), 2, n()))
)
}
OR.cepha.strata.C3G.n=cepha.strata.C3G %>% filter(Year.ID %in% subset(d1016.strata.C3G,Case==1)$Year.ID) %>% group_by(Drug) %>% tally(sort=T) %>% left_join(
OR.cepha.strata.C3G , by = c("Drug" = "rowname")
)
OR.cepha.strata.C3G.n %>% print()
1
2
3
4
5
6
7
8
9
10
11
12
## # A tibble: 9 x 5
## Drug n OR lower upper
## <chr> <int> <dbl> <dbl> <dbl>
## 1 cefotaxim 557 0.90 0.795 1.02
## 2 ceftriaxon 307 1.07 0.931 1.24
## 3 ceftazidim 252 1.20 1.027 1.40
## 4 cefoperazon 88 1.26 1.017 1.56
## 5 cefoperazon.sulbactam 31 1.36 0.894 2.03
## 6 ceftizoxim 28 1.00 0.649 1.49
## 7 cefixim 10 0.25 0.122 0.44
## 8 cefpodoxim 5 0.36 0.125 0.81
## 9 cefdinir 2 0.51 0.081 1.78
1
2
3
4
5
6
7
8
9
OR.cepha.strata.C3G.n %>% unique() %>% mutate(Drug=paste0(Drug,'(n=',n,")")) %>%
arrange(n) %>% mutate(Drug = factor(Drug, Drug)) %>%
ggplot( aes(x=Drug, y=OR, ymin=lower, ymax=upper)) +
geom_errorbar() +
geom_point(aes(shape=if_else(lower>1,T,F))) + #,col="blue",shape=5
geom_hline(yintercept=1, lty=2) + # add a dotted line at x=1 after flip
coord_flip() + # flip coordinates (puts labels on y axis)
xlab("Drug") + ylab("Adjusted ROR (95% CI)") +
theme_bw() # use a white background

1
2
3
4
5
6
7
#
# OR.cepha.strata.C3G.n %>%
# arrange(desc(n)) %>% mutate(OR=round(OR,2),
# lower=round(lower,2),
# upper=round(upper,2)) %>%
#
# write.csv2("OR.cepha.strata.C3G.n.csv",row.names=F)
6. Risk relatifs between C3G with cefotaxim as refrence
1
2
3
4
5
## Réordonnancement de d1016.strata.C3G$C3G
d1016.strata.C3G$C3G <- factor(d1016.strata.C3G$C3G, levels=c("cefotaxim", "ceftriaxon", "ceftazidim", "cefixim", "cefoperazon", "ceftizoxim", "cefpodoxim", "cefoperazon.sulbactam", "cefdinir", "cefodoxim", "cefetamet", "ceftriaxon.sulbactam"))
d1016.strata.C3G %>% group_by(C3G) %>% tally(sort=T)
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
## # A tibble: 12 x 2
## C3G n
## <fctr> <int>
## 1 cefotaxim 3115
## 2 ceftriaxon 1561
## 3 ceftazidim 1192
## 4 cefoperazon 405
## 5 cefixim 181
## 6 ceftizoxim 149
## 7 cefoperazon.sulbactam 130
## 8 cefpodoxim 65
## 9 cefdinir 19
## 10 ceftriaxon.sulbactam 4
## 11 cefodoxim 1
## 12 cefetamet 1
1
2
3
4
5
6
7
8
9
10
11
12
d1016.strata.C3G %>% select(Year,Age,Sex,Case,C3G) %>%
mutate(Case=as.factor(Case)) %>%
glm(formula=Case~Year+Age+Sex+C3G,family=binomial(link="logit")) %>% OR95()%>%
tibble::rownames_to_column() %>% filter(between(row_number(), 9, n())) %>%
ggplot( aes(x=rowname, y=OR, ymin=lower, ymax=upper)) +
geom_errorbar() +
geom_point(aes(shape=if_else(lower>1,T,F))) + #,col="blue",shape=5
geom_hline(yintercept=1, lty=2) + # add a dotted line at x=1 after flip
coord_flip(ylim=c(0,3)) + # flip coordinates (puts labels on y axis)
xlab("C3G diferent risks") + ylab("Adjusted ROR (95% CI)") +
theme_bw() # use a white background
